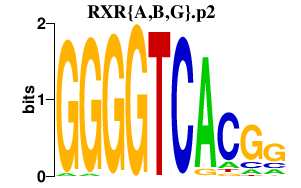
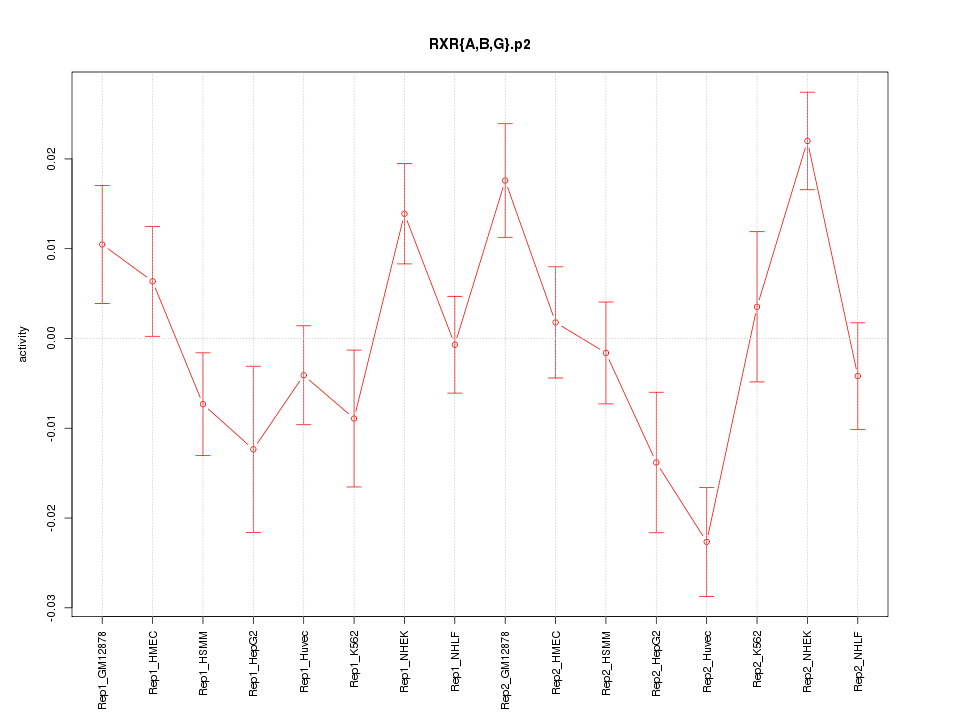
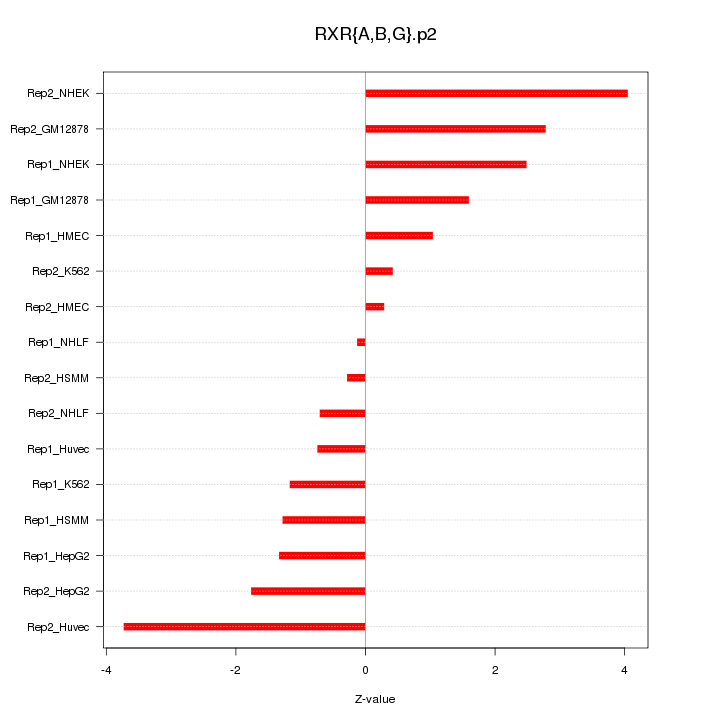
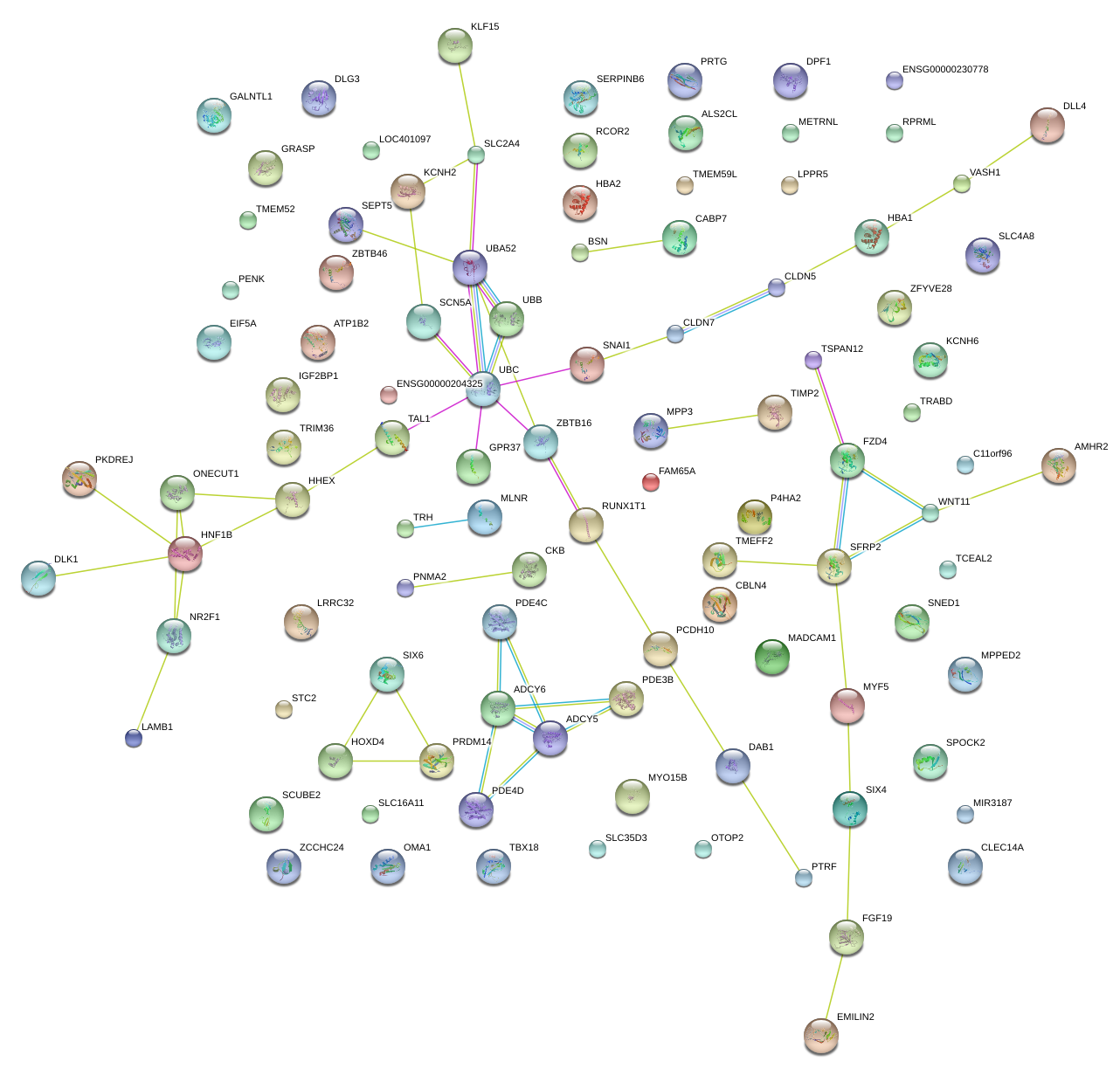
|
chr17_+_44429748
|
5.073
|
NM_001160423
NM_006546
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1
|
|
chr14_-_103058568
|
2.241
|
|
CKB
|
creatine kinase, brain
|
|
chr12_+_50686990
|
2.066
|
NM_181711
|
GRASP
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein
|
|
chr15_-_53822468
|
1.729
|
NM_173814
|
PRTG
|
protogenin
|
|
chr5_-_131591380
|
1.659
|
NM_001017974
NM_001142598
NM_001142599
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II
|
|
chr5_-_172688024
|
1.648
|
|
STC2
|
stanniocalcin 2
|
|
chr8_-_71146103
|
1.636
|
NM_024504
|
PRDM14
|
PR domain containing 14
|
|
chr4_+_134292630
|
1.616
|
|
PCDH10
|
protocadherin 10
|
|
chr11_-_75595098
|
1.588
|
NM_004626
|
WNT11
|
wingless-type MMTV integration site family, member 11
|
|
chr8_-_71146022
|
1.554
|
|
PRDM14
|
PR domain containing 14
|
|
chr17_+_7125702
|
1.499
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr3_-_124650081
|
1.486
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr22_+_18081968
|
1.437
|
NM_002688
|
SEPT5
|
septin 5
|
|
chr3_+_49566901
|
1.363
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr5_-_172687798
|
1.352
|
|
STC2
|
stanniocalcin 2
|
|
chr22_+_48966394
|
1.247
|
|
TRABD
|
TraB domain containing
|
|
chr20_-_61933012
|
1.234
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr12_-_50502474
|
1.200
|
NM_001013690
|
FIGNL2
|
fidgetin-like 2
|
|
chr4_+_134293171
|
1.170
|
|
PCDH10
|
protocadherin 10
|
|
chr22_+_48966480
|
1.137
|
NM_025204
|
TRABD
|
TraB domain containing
|
|
chr17_+_70431964
|
1.110
|
NM_178160
|
OTOP2
|
otopetrin 2
|
|
chr14_+_100270222
|
1.095
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr22_-_45037882
|
1.092
|
NM_006071
|
PKDREJ
|
polycystic kidney disease (polycystin) and REJ homolog (sperm receptor for egg jelly homolog, sea urchin)
|
|
chr22_+_18085990
|
1.088
|
|
SEPT5-GP1BB
SEPT5
|
SEPT5-GP1BB read-through transcript
septin 5
|
|
chr17_-_33178987
|
1.087
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr22_+_18085891
|
1.072
|
|
SEPT5
|
septin 5
|
|
chr17_-_6887965
|
1.049
|
NM_153357
|
SLC16A11
|
solute carrier family 16, member 11 (monocarboxylic acid transporter 11)
|
|
chr17_+_71097147
|
1.043
|
|
MYO15B
|
myosin XVB pseudogene
|
|
chr11_-_76059435
|
1.028
|
NM_001128922
|
LRRC32
|
leucine rich repeat containing 32
|
|
chr17_-_33179118
|
1.017
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr5_+_92946348
|
1.011
|
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr1_+_14797764
|
0.999
|
NM_015209
NM_201628
|
KAZN
|
kazrin, periplakin interacting protein
|
|
chr11_-_86343859
|
0.973
|
NM_012193
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr11_+_113436438
|
0.966
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr11_+_43920385
|
0.954
|
NM_001145033
|
C11orf96
|
chromosome 11 open reading frame 96
|
|
chr15_+_39008822
|
0.949
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr11_+_14621972
|
0.945
|
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr11_-_63440891
|
0.922
|
NM_173587
|
RCOR2
|
REST corepressor 2
|
|
chrX_+_69591639
|
0.922
|
NM_001166278
|
DLG3
|
discs, large homolog 3 (Drosophila)
|
|
chr19_+_447453
|
0.915
|
NM_130760
NM_130762
|
MADCAM1
|
mucosal vascular addressin cell adhesion molecule 1
|
|
chr5_-_59225377
|
0.915
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_74432671
|
0.892
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr17_-_74433048
|
0.883
|
NM_003255
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr7_-_124192265
|
0.882
|
|
GPR37
|
G protein-coupled receptor 37 (endothelin receptor type B-like)
|
|
chr12_+_79634820
|
0.865
|
NM_005593
|
MYF5
|
myogenic factor 5
|
|
chr1_-_99242765
|
0.862
|
NM_001010861
NM_001037317
|
LPPR5
|
lipid phosphate phosphatase-related protein type 5
|
|
chr3_+_129690735
|
0.860
|
|
|
|
|
chr6_+_137285071
|
0.855
|
NM_001008783
|
SLC35D3
|
solute carrier family 35, member D3
|
|
chr5_-_114543632
|
0.845
|
NM_001017397
NM_001017398
NM_018700
|
TRIM36
|
tripartite motif containing 36
|
|
chr22_+_28446288
|
0.812
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr4_-_2336328
|
0.807
|
NM_001172659
NM_001172660
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr14_+_68796621
|
0.796
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr11_-_69228286
|
0.792
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr8_-_57521702
|
0.792
|
NM_001135690
|
PENK
|
proenkephalin
|
|
chr18_+_2896956
|
0.790
|
|
EMILIN2
|
elastin microfibril interfacer 2
|
|
chr19_-_43412170
|
0.787
|
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr17_+_78630793
|
0.774
|
NM_001004431
|
METRNL
|
meteorin, glial cell differentiation regulator-like
|
|
chr3_+_161426322
|
0.769
|
|
LOC401097
|
hypothetical protein LOC401097
|
|
chr10_+_94439658
|
0.763
|
NM_002729
|
HHEX
|
hematopoietically expressed homeobox
|
|
chr15_-_50869379
|
0.762
|
NM_004498
|
ONECUT1
|
one cut homeobox 1
|
|
chr19_-_43412152
|
0.761
|
NM_001135156
|
DPF1
|
D4, zinc and double PHD fingers family 1
|
|
chr8_-_93184629
|
0.757
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_192767494
|
0.753
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr15_-_38362078
|
0.747
|
NM_001190479
|
LOC100131244
|
hypothetical protein LOC100131244
|
|
chr8_-_26427304
|
0.738
|
NM_007257
|
PNMA2
|
paraneoplastic antigen MA2
|
|
chr2_+_176724358
|
0.731
|
NM_014621
|
HOXD4
|
homeobox D4
|
|
chr3_+_161425946
|
0.728
|
NM_001168214
|
LOC401097
|
hypothetical protein LOC401097
|
|
chr3_-_127558747
|
0.728
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr19_+_18584666
|
0.727
|
NM_012109
|
TMEM59L
|
transmembrane protein 59-like
|
|
chr19_-_772913
|
0.724
|
NM_024888
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr14_+_60045621
|
0.722
|
NM_007374
|
SIX6
|
SIX homeobox 6
|
|
chr22_-_17892001
|
0.717
|
|
CLDN5
|
claudin 5
|
|
chr17_-_37828643
|
0.709
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr11_-_86343737
|
0.698
|
|
FZD4
|
frizzled homolog 4 (Drosophila)
|
|
chr19_-_772966
|
0.695
|
|
LPPR3
|
lipid phosphate phosphatase-related protein type 3
|
|
chr3_-_38666122
|
0.695
|
NM_000335
NM_001099404
NM_001099405
NM_198056
NM_001160160
NM_001160161
|
SCN5A
|
sodium channel, voltage-gated, type V, alpha subunit
|
|
chr19_-_18198234
|
0.690
|
NM_001098818
|
PDE4C
|
phosphodiesterase 4C, cAMP-specific
|
|
chr22_-_17891777
|
0.690
|
|
CLDN5
|
claudin 5
|
|
chr7_-_120285245
|
0.687
|
|
TSPAN12
|
tetraspanin 12
|
|
chr17_-_74432800
|
0.684
|
|
TIMP2
|
TIMP metallopeptidase inhibitor 2
|
|
chr13_+_48692472
|
0.672
|
NM_001507
|
MLNR
|
motilin receptor
|
|
chr3_+_131175803
|
0.671
|
NM_007117
|
TRH
|
thyrotropin-releasing hormone
|
|
chr10_-_73518269
|
0.670
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr14_-_37795001
|
0.667
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr17_+_7495436
|
0.655
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr2_+_241586927
|
0.653
|
NM_001080437
|
SNED1
|
sushi, nidogen and EGF-like domains 1
|
|
chr19_+_18543613
|
0.653
|
NM_001033930
NM_003333
|
UBA52
|
ubiquitin A-52 residue ribosomal protein fusion product 1
|
|
chr1_-_57661373
|
0.651
|
|
DAB1
|
disabled homolog 1 (Drosophila)
|
|
chr1_-_47468029
|
0.649
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr11_-_9069668
|
0.647
|
NM_001170690
NM_020974
|
SCUBE2
|
signal peptide, CUB domain, EGF-like 2
|
|
chr14_+_76298693
|
0.641
|
|
VASH1
|
vasohibin 1
|
|
chr10_-_73518284
|
0.625
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr1_-_1840564
|
0.624
|
NM_178545
|
TMEM52
|
transmembrane protein 52
|
|
chrX_+_101267315
|
0.624
|
NM_080390
|
TCEAL2
|
transcription elongation factor A (SII)-like 2
|
|
chr17_-_76985538
|
0.622
|
|
|
|
|
chr16_+_66129481
|
0.621
|
|
FAM65A
|
family with sequence similarity 65, member A
|
|
chr20_+_48032918
|
0.618
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr17_-_7106497
|
0.617
|
|
CLDN7
|
claudin 7
|
|
chr10_-_80875097
|
0.617
|
NM_153367
|
ZCCHC24
|
zinc finger, CCHC domain containing 24
|
|
chr6_-_85529031
|
0.616
|
|
TBX18
|
T-box 18
|
|
chr2_-_192767844
|
0.614
|
NM_016192
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr17_-_42411587
|
0.613
|
NM_203400
|
RPRML
|
reprimo-like
|
|
chr12_+_50104859
|
0.613
|
NM_001039960
NM_004858
|
SLC4A8
|
solute carrier family 4, sodium bicarbonate cotransporter, member 8
|
|
chr6_-_2916280
|
0.607
|
|
SERPINB6
|
serpin peptidase inhibitor, clade B (ovalbumin), member 6
|
|
chr4_-_154929657
|
0.607
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|
|
chr7_-_150283794
|
0.607
|
NM_172057
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr7_-_107430566
|
0.606
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr11_-_30564338
|
0.605
|
NM_001145399
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr14_-_60260211
|
0.604
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr20_-_54013418
|
0.602
|
NM_080617
|
CBLN4
|
cerebellin 4 precursor
|
|
chr1_+_27192719
|
0.599
|
NM_001013642
|
TRNP1
|
TMF1-regulated nuclear protein 1
|
|
chr12_+_52105131
|
0.598
|
|
AMHR2
|
anti-Mullerian hormone receptor, type II
|
|
chr3_-_46710137
|
0.592
|
NM_001190707
NM_147129
|
ALS2CL
|
ALS2 C-terminal like
|
|
chr17_+_7151653
|
0.592
|
|
EIF5A
|
eukaryotic translation initiation factor 5A
|
|
chr16_+_162845
|
0.585
|
NM_000517
|
HBA1
HBA2
|
hemoglobin, alpha 1
hemoglobin, alpha 2
|
|
chrX_-_138114100
|
0.583
|
|
FGF13
|
fibroblast growth factor 13
|
|
chr17_-_7106987
|
0.579
|
NM_001185023
NM_001307
|
CLDN7
|
claudin 7
|
|
chr11_+_817584
|
0.574
|
NM_173584
|
EFCAB4A
|
EF-hand calcium binding domain 4A
|
|
chrX_+_136476353
|
0.572
|
|
ZIC3
|
Zic family member 3 (odd-paired homolog, Drosophila)
|
|
chr19_+_14982538
|
0.570
|
NM_173482
|
CCDC105
|
coiled-coil domain containing 105
|
|
chr3_-_51968995
|
0.567
|
|
PCBP4
|
poly(rC) binding protein 4
|
|
chr2_-_228754585
|
0.567
|
NM_001142644
NM_030623
|
SPHKAP
|
SPHK1 interactor, AKAP domain containing
|
|
chr7_-_126679540
|
0.566
|
NM_001127323
|
GRM8
|
glutamate receptor, metabotropic 8
|
|
chr14_+_28306633
|
0.566
|
|
FOXG1
|
forkhead box G1
|
|
chr20_+_20296744
|
0.566
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chrX_-_44945025
|
0.565
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr7_+_78920846
|
0.559
|
|
LOC100505881
|
hypothetical LOC100505881
|
|
chr20_-_3097070
|
0.553
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr4_+_2031036
|
0.551
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr19_-_15421761
|
0.550
|
NM_021241
|
WIZ
|
widely interspaced zinc finger motifs
|
|
chr10_-_17536181
|
0.550
|
NM_001004470
|
ST8SIA6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6
|
|
chr17_+_26742767
|
0.550
|
NM_032932
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr7_+_94374862
|
0.547
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr2_-_27457088
|
0.545
|
NM_144631
|
ZNF513
|
zinc finger protein 513
|
|
chr10_-_134605978
|
0.543
|
NM_173572
|
C10orf93
|
chromosome 10 open reading frame 93
|
|
chr20_+_44091219
|
0.537
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr6_+_21774624
|
0.533
|
|
FLJ22536
|
hypothetical locus LOC401237
|
|
chr22_+_43451351
|
0.533
|
NM_001017530
NM_001198721
NM_015366
|
PRR5
|
proline rich 5 (renal)
|
|
chr5_-_92942680
|
0.532
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr1_-_86394695
|
0.531
|
NM_152890
|
COL24A1
|
collagen, type XXIV, alpha 1
|
|
chr1_-_26245031
|
0.529
|
NM_001004434
NM_032513
|
SLC30A2
|
solute carrier family 30 (zinc transporter), member 2
|
|
chr19_+_54314292
|
0.529
|
NM_003660
|
PPFIA3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr14_+_100263028
|
0.528
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chr5_+_149526533
|
0.525
|
NM_001804
|
CDX1
|
caudal type homeobox 1
|
|
chr17_+_63609317
|
0.524
|
|
LOC100499466
|
hypothetical LOC100499466
|
|
chr7_-_150306169
|
0.524
|
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr20_+_3000265
|
0.521
|
NM_000915
|
OXT
|
oxytocin, prepropeptide
|
|
chr20_+_57612996
|
0.518
|
NM_080672
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr12_+_50587468
|
0.518
|
NM_000020
|
ACVRL1
|
activin A receptor type II-like 1
|
|
chr1_+_43769065
|
0.517
|
NM_002840
NM_130440
|
PTPRF
|
protein tyrosine phosphatase, receptor type, F
|
|
chr17_+_56831951
|
0.515
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr1_+_12149662
|
0.515
|
|
TNFRSF1B
|
tumor necrosis factor receptor superfamily, member 1B
|
|
chr14_-_76807302
|
0.512
|
NM_021257
|
NGB
|
neuroglobin
|
|
chr7_-_78920806
|
0.510
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr9_+_102275310
|
0.507
|
NM_003692
|
TMEFF1
|
transmembrane protein with EGF-like and two follistatin-like domains 1
|
|
chr6_+_96570565
|
0.507
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr9_-_70818829
|
0.507
|
NM_002732
|
PRKACG
|
protein kinase, cAMP-dependent, catalytic, gamma
|
|
chr19_-_7244876
|
0.506
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chrX_+_38305634
|
0.505
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr7_-_27191287
|
0.505
|
NM_005523
|
HOXA11
|
homeobox A11
|
|
chrX_-_30237403
|
0.504
|
NM_000475
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1
|
|
chr4_-_86106567
|
0.504
|
NM_014991
|
WDFY3
|
WD repeat and FYVE domain containing 3
|
|
chr22_+_16973556
|
0.503
|
|
TUBA8
|
tubulin, alpha 8
|
|
chr19_-_56260135
|
0.503
|
NM_015596
|
KLK13
|
kallikrein-related peptidase 13
|
|
chr6_+_69401660
|
0.502
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr19_-_18867868
|
0.501
|
NM_021267
NM_198207
NM_001492
|
LASS1
GDF1
|
LAG1 homolog, ceramide synthase 1
growth differentiation factor 1
|
|
chr17_+_26742903
|
0.499
|
|
RAB11FIP4
|
RAB11 family interacting protein 4 (class II)
|
|
chr14_-_103058865
|
0.497
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr9_-_138772506
|
0.496
|
NM_178469
|
LCN8
|
lipocalin 8
|
|
chr8_+_26427378
|
0.494
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr19_+_44697592
|
0.491
|
NM_182704
|
SELV
|
selenoprotein V
|
|
chr8_+_69027166
|
0.490
|
|
PREX2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2
|
|
chr10_-_135232865
|
0.490
|
NM_130784
|
SYCE1
|
synaptonemal complex central element protein 1
|
|
chr18_+_75261231
|
0.489
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr8_+_22479115
|
0.488
|
NM_001018003
|
SORBS3
|
sorbin and SH3 domain containing 3
|
|
chr1_-_37272295
|
0.488
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr7_+_44110527
|
0.487
|
|
AEBP1
|
AE binding protein 1
|
|
chr22_+_36531059
|
0.483
|
NM_005318
|
H1F0
|
H1 histone family, member 0
|
|
chr17_-_45562112
|
0.481
|
NM_174920
|
SAMD14
|
sterile alpha motif domain containing 14
|
|
chr2_+_104838177
|
0.480
|
NM_006236
|
POU3F3
|
POU class 3 homeobox 3
|
|
chrX_-_47394808
|
0.479
|
|
ELK1
|
ELK1, member of ETS oncogene family
|
|
chr20_+_44091265
|
0.479
|
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr15_+_30797466
|
0.477
|
NM_001191322
NM_001191323
NM_013372
|
GREM1
|
gremlin 1
|
|
chr17_-_4967120
|
0.477
|
NM_014519
|
ZNF232
|
zinc finger protein 232
|
|
chr14_-_73962557
|
0.475
|
NM_001105579
|
TMEM90A
|
transmembrane protein 90A
|
|
chr19_-_646460
|
0.474
|
NM_214710
|
PRSS57
|
protease, serine, 57
|
|
chr4_+_996251
|
0.474
|
NM_021923
|
FGFRL1
|
fibroblast growth factor receptor-like 1
|
|
chr10_-_73518315
|
0.473
|
|
SPOCK2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2
|
|
chr7_+_44110470
|
0.472
|
NM_001129
|
AEBP1
|
AE binding protein 1
|
|
chr7_-_94123308
|
0.472
|
|
SGCE
|
sarcoglycan, epsilon
|
|
chr10_+_26545241
|
0.470
|
NM_000818
NM_001134366
|
GAD2
|
glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa)
|
|
chr4_-_13155135
|
0.470
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr2_+_176724583
|
0.470
|
|
|
|
|
chr14_-_37795290
|
0.469
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr5_+_169465478
|
0.468
|
NM_012188
NM_144769
|
FOXI1
|
forkhead box I1
|
|
chr17_+_37222731
|
0.466
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr1_-_38285033
|
0.464
|
NM_002699
|
POU3F1
|
POU class 3 homeobox 1
|
|
chr17_+_37222726
|
0.463
|
|
FKBP10
|
FK506 binding protein 10, 65 kDa
|
|
chr4_-_41848874
|
0.462
|
|
BEND4
|
BEN domain containing 4
|
|
chr9_-_16860719
|
0.460
|
NM_017637
|
BNC2
|
basonuclin 2
|